We discovered that decomposing manipulation trajectories into object alignment followed by object interaction enables learning new tasks from single demonstrations, compared to tens or hundreds typically required. Through 3,450 real-world experiments, we developed Multi-Task Trajectory Transfer (MT3), which combines trajectory decomposition with retrieval in learned latent spaces. Thanks to MT3's data efficiency, we were able to teach a robot 1000 distinct manipulation tasks in less than 24 hours of human demonstrator time, while also generalizing effectively to novel objects. This work was featured on the cover of the November 2025 issue of Science Robotics.
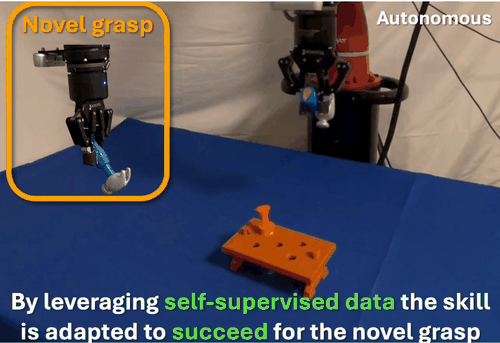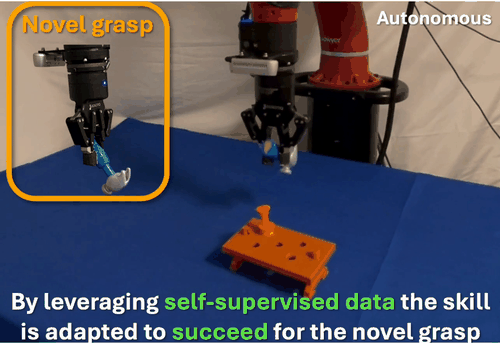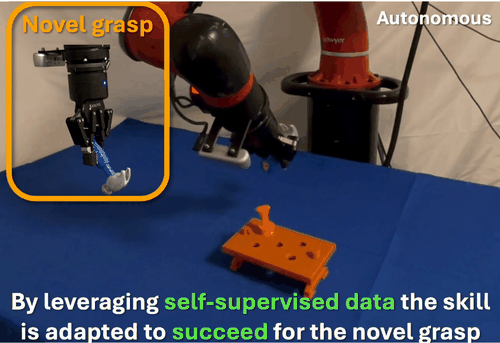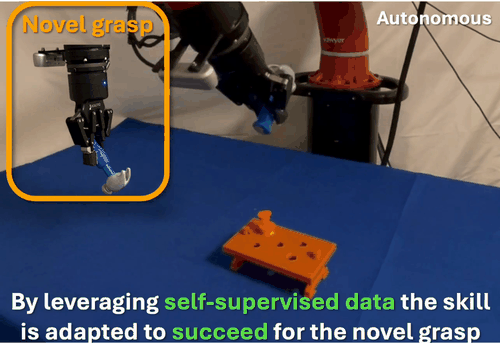
We found that robots typically fail to transfer skills when grasping objects differently than during training. To address this, we developed a self-supervised data collection method that enables grasp-invariant skill transfer. Our approach allows robots to successfully adapt manipulation skills to novel grasps without requiring additional human demonstrations, demonstrating robust generalization across varying grasp configurations through systematic real-world validation.
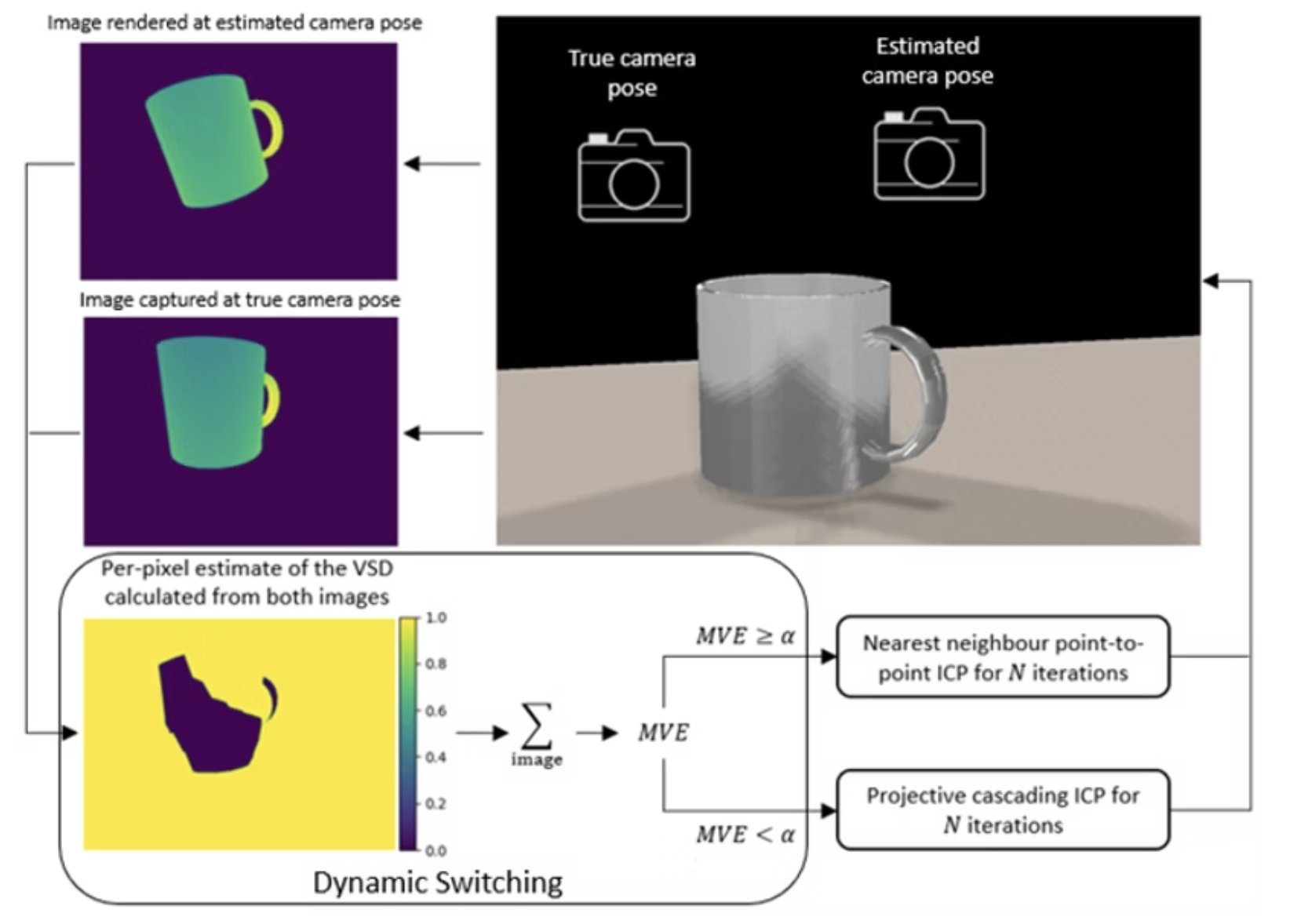
We investigated how combining trajectory transfer with unseen object pose estimation enables robots to learn new tasks from single demonstrations, eliminating the need for additional data collection or training. Through systematic experimentation, we characterized the impact of pose estimation errors and camera calibration errors on task success rates, revealing critical tolerance thresholds for reliable one-shot learning. This analysis provides practical insights for deploying vision-based manipulation systems in real-world environments.